Serotonin Docking - Demo Pipeline
Welcome to xyna.bio!
Here we guide you through a demo pipeline, showcasing nodes, features and functions on the example of docking Serotonin to a human Serotonin Receptor.
File Upload
We start by downloading the FASTA for the Serotonin Receptor 6 (can be found here: https://www.ncbi.nlm.nih.gov/protein/P50406.1?report=fasta). To upload your file, open the "Files" page in the left menu tab and click on the "Upload" button. After successful upload you will be notified and should now see your file in the file list.
Pipeline Setup
-
Click right to open the menu and search through our node library. Search and select the first node: Load Fasta and search for your Serotonin Fasta file to load.
-
Select OmegaFold as your second node. Connect the pipeline by dragging the connection from output (here: "Sequence" of the Load Fasta node) to the input (here: "Input protein sequence" of the Omega Fold node). The loaded sequence then gets forwarded to the OmegaFold node, where its 3D structure gets predicted.
-
Select the Protein Structure Viewer node, and connect it to the "predicted strucutre" output to compute a preview of the structure.
Click on "Open in MolStar" to automatically open your protein strucutre in our internal MolStar visualization suite. There you can take a closer look into the 3D structure!
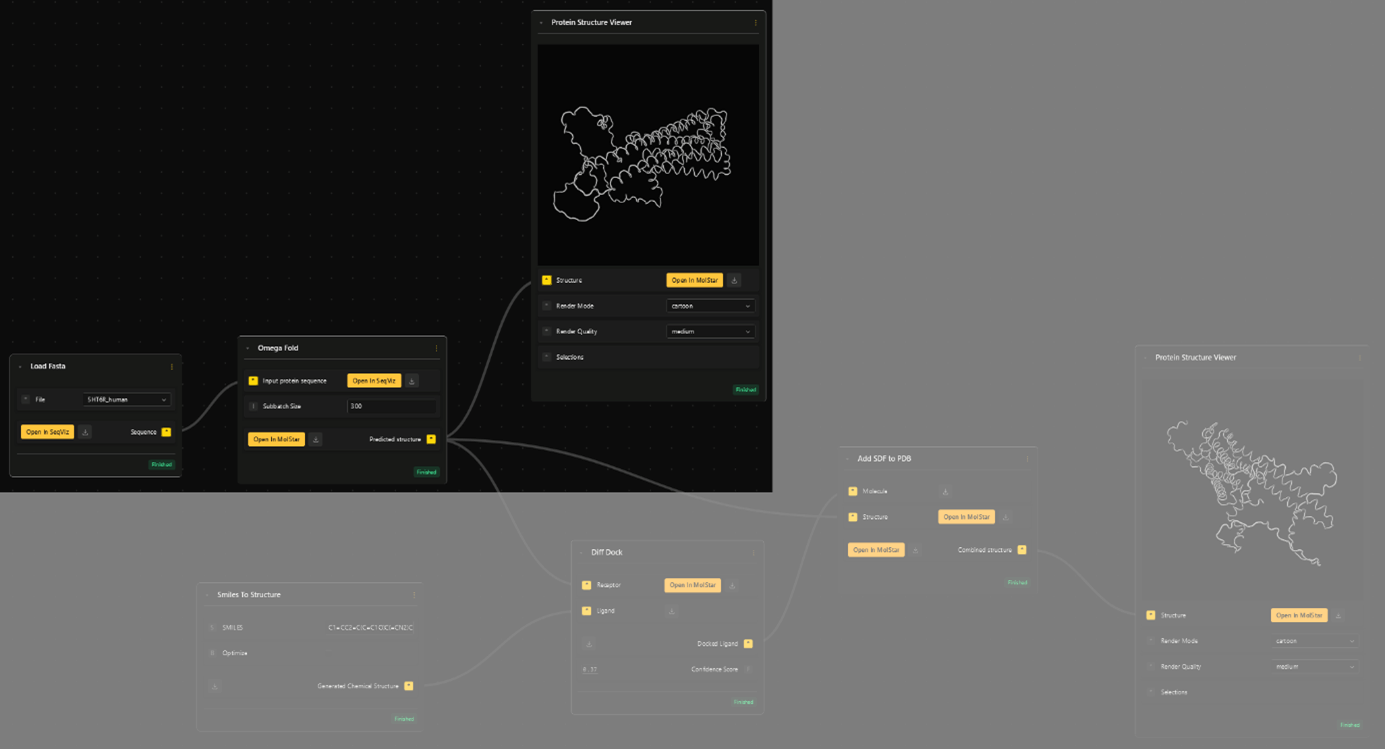
- To do the docking, we pipe the predicted structure into the "Receptor" input of the DiffDock node.
The second input, "Ligand" for DiffDock is the chemical structure of the serotonin molecule. For that we need the SMILES (Simplified Molecular-Input Line-Entry System) string of the serotonin molecule, which is easily found online.
- Select the Smiles to Structure node. Copy the string into the input field. This node will transform the information of the molecule from a SMILES to SDF (Spatial Data File) format, which is needed for downstream docking.
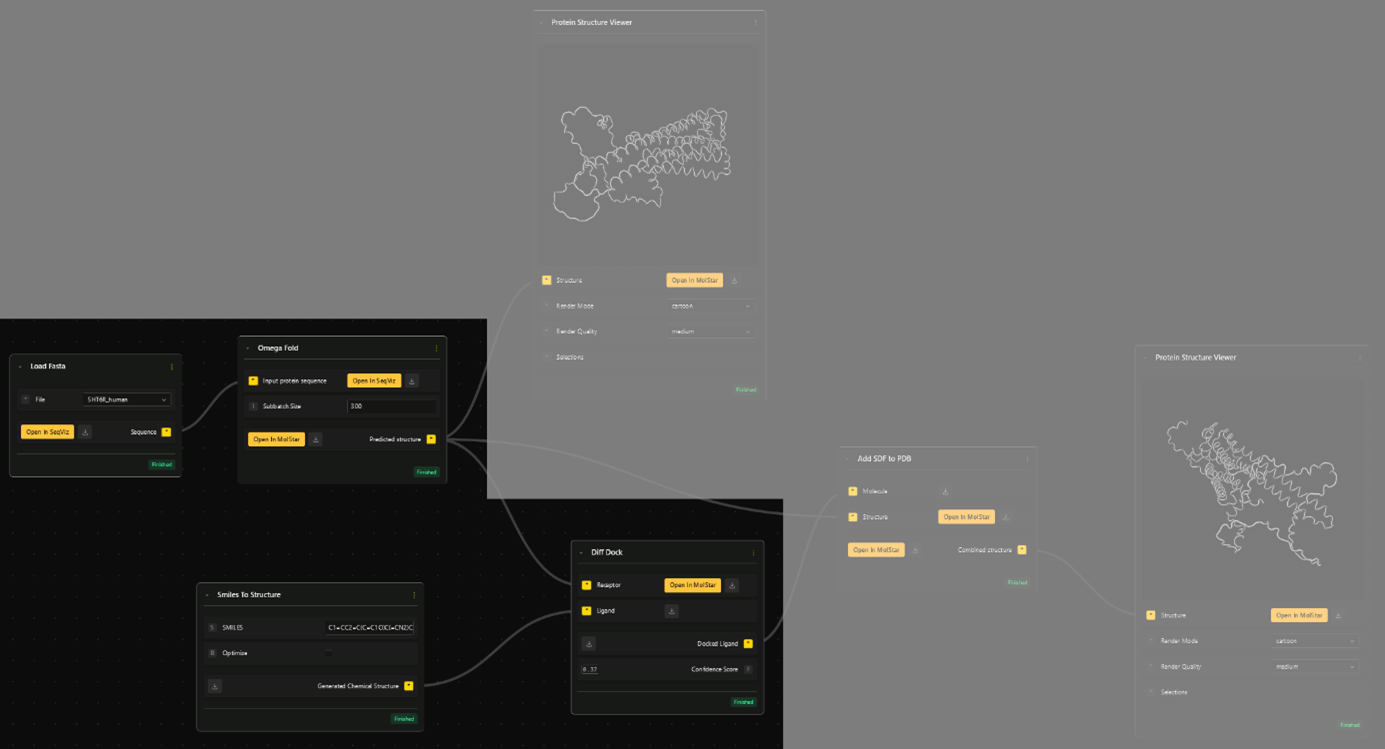
Diff Dock outputs the spatial information for the docked molecule. To see the molecule in its docked position, together with the folded receptor, need combine the structures with the Add SDF to PDB node.
- Select the Add SDF to PDB node and input the "Docked Ligand" (Diff Dock) output as "Molecule" and "Predicted structure" (Omega Fold) as "Structure".
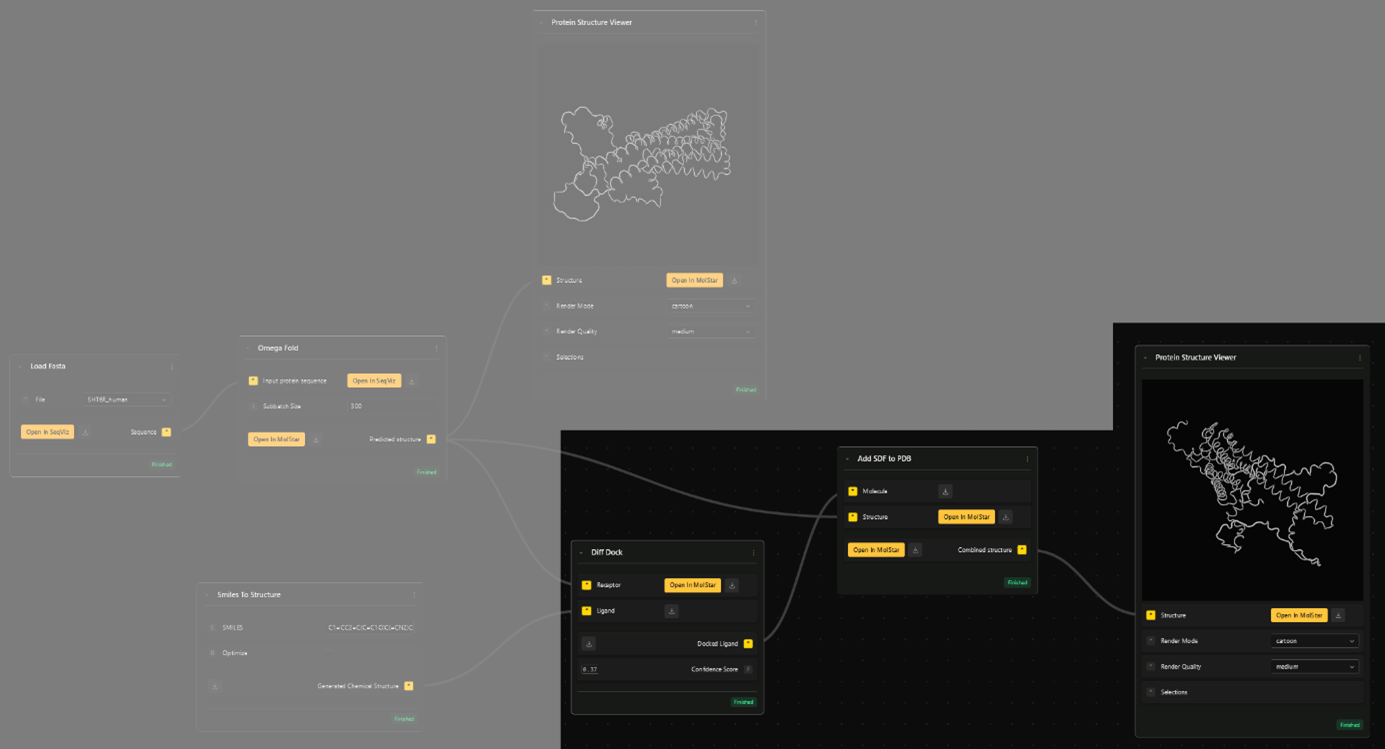
- The combined structure can then be viewed with another "Protein Structure Viewer". To get an even better view, click the "Open in MolStar" button to enter the visualization environment.
Bonus Node
- To quantify the binding, select the Protein Ligand Binding Affinity node and connect the input field with the "combined structure" (Add SDF to PDB).