Bulk RNA-seq
Xyna.bio combines a comprehensive suite of tools for bulk RNA-seq data analysis, providing a streamlined platform for setting up tailored pipelines. The node based modular design allows users to configure and run each step of the analysis efficiently, from quality control to differential expression analysis. As bulk RNA-seq is a varied field with many possible applications and, as such, many possible pipelines there is no one "complete" pipeline or even a set of absolutely necessary nodes. Because of this below, we present an overview of all the pipeline's integrated nodes and their possible applications. In addition there are figures showing an extensive differential expression analysis pipeline, that integrates most of the nodes and can be seen as a genral purpose example.
The end of a pipeline usually consists of a MultiQC node that takes results generated at several stages of the pipeline and assembles them into one report that provides an overview containing most of the information necessary to interpret the sequencing results.
The pipeline consists of four main steps:
1. Pre-Processing
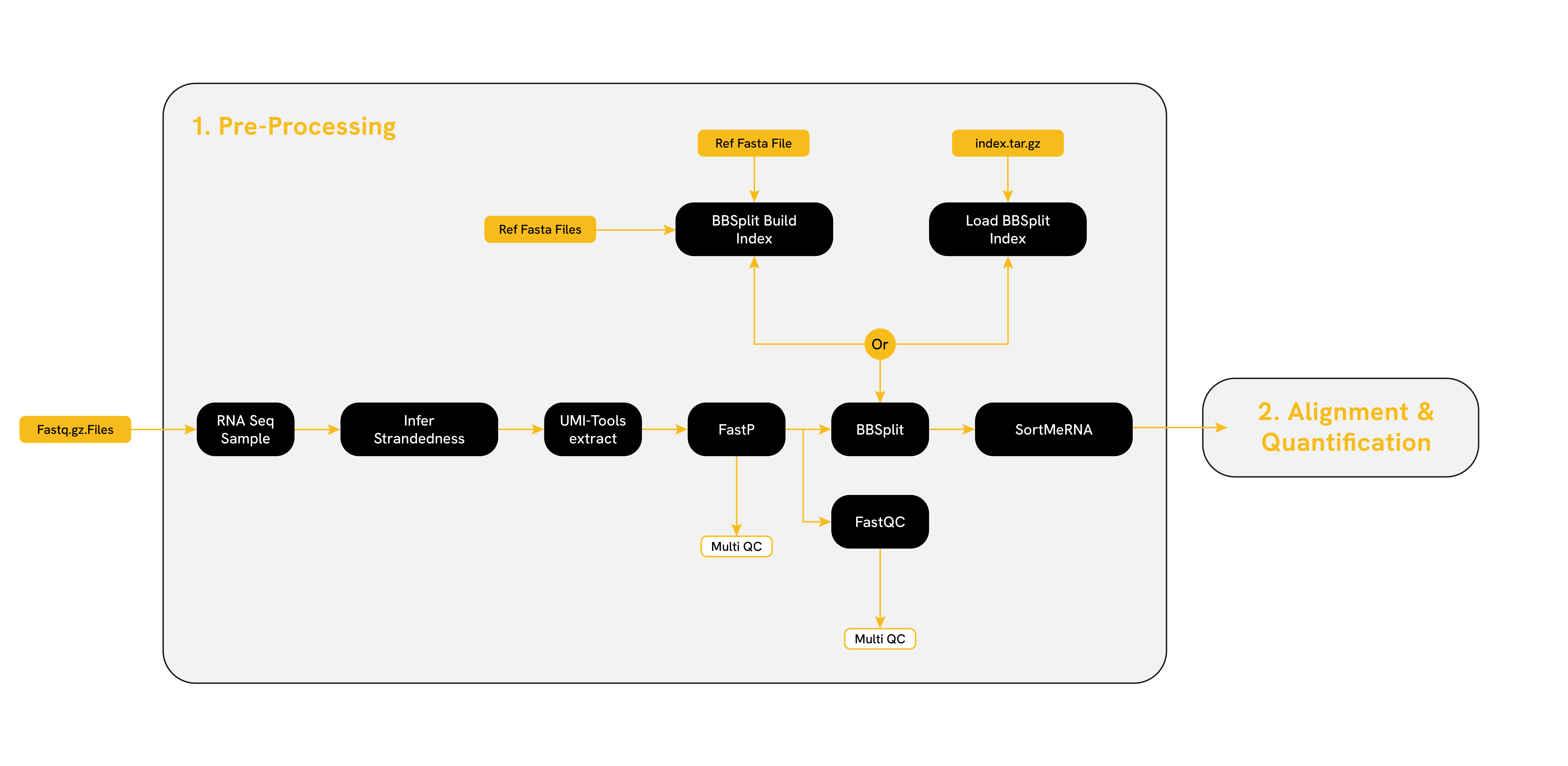
Pre-processing data ensures starting the analysis with only high quality reads and removes artifacts that were produced by the process of RNA sequencing. It includes extraction of UMIs (unique molecular identifiers) that were inserted during PCR (UMI-Tools extract), removal of adapter sequences + filtering of short and low quality reads (FastP) as well as extraction of ribosomal RNA (SortMeRNA). Further, strandedness of the reads is inferred (Infer Strandedness) and mapping to one or multiple references can be done (BBSplit). Additionally, several quality metrics are computed for an extensive quality control in the end (FastQC).
RNA Seq Sample
Loads a .fastq.gz file containing RNA-seq reads. These unprocessed reads are the basis on which the analysis of the pipeline will be performed.
Input:
This node shows a selection of .fastq.gz file from the files section that were uploaded there previously. There are public sources for bulk RNA-seq datasets like the Gene Expression Omnibus that can be used as a starting point.
Output:
- Samples: Reference to one or multiple .fastq.gz files for further processing steps.
Infer Strandedness
Infers the strandedness for fastq files and adds it as metadata to the samples. The strandedness of a dataset usually comes from the way the reads were generated and is important to know for better analysis downstream.
Tool used: Salmon
Input:
-
Samples: List of fastq RNA-seq samples without strandedness metadata.
-
Subsamples: List of subsamples of the above 'Samples'. The samples and subsamples are matched based on their name. This will speed up the inference time, because fewer reads are handled.
-
Salmon Index: Genome index matching the organism the reads were obtained from.
Input Parameters:
-
Stranded Threshold: The portion of reads that have to match a certain strandedness to classify the reads as a whole as that strandedness.
-
Unstranded Threshold: The portion of reads that have different strandedness for the reads to be classified as unstranded.
Output:
- Stranded Samples: List of RNA-seq samples with strandedness metadata (Fastq). The strandedness of a sample can be
forward,reverseorunstranded.
UMI-Tools extract
Extracts Unique Molecular Identifiers (UMI) from the reads. This will recognize UMIs with a given pattern, remove them from the read and add the UMI to the name of the read so it can be identified later.
Tool used: UMI-tools
Input:
- Samples: List of fastq RNA-seq samples with UMIs present in the reads.
Input Parameters:
-
Extract Method: Method to specify the pattern by which to recognize the UMIs. For a detailed breakdown on how to set the right pattern see the official documentation.
-
Barcode 1: Barcode to use on the first reads.
-
Barcode 2: Barcode to use on the second reads.
-
Seperator: The seperator to use to add the UMIs to the read name.
Output:
-
Processed Samples: List of RNA-seq Samples with UMIs extracted and added to the read name (fastq).
-
Logs: Log file describing the run for each Sample.
FastP
FastP processes and does quality control on Fastq data. This includes filtering short and low quality reads, trimming adapter sequences and generating quality reports in HTML and JSON formats. These reports are usually fed into MultiQC to assemble a combined report.
Tool used: FastP
Input:
- Samples: List of unprocessed fastq RNA-seq samples.
Input Parameters:
-
Adapter Fasta: (optional) Fasta file containing the adapter sequence added to the reads for sequencing. This sequence can also be detected automatically, but for single end reads it should still be specified as the detection may be inaccurate. for more information on how to setup an adapter fasta file see here
-
Minimum Reads: The minimum number of reads that have to be left in a sample after filtering. If the sample contains less reads then it is discarded and will not be used in the downstread analysis. Reports, logs, merged and failed files are still generated.
-
Enable Merging: If set this will generate a file where overlapping paired end reads are combined into a single read.
-
Save Discarded Reads: If set this will generate a file containing all the reads that were filtered out by FastP.
Output:
-
Filtered Samples: List of trimmed and filtered RNA-seq samples (fastq).
-
Merge Fastq: (optional) List of merged reads.
-
Discarded Fastq: (optional) List of discarded reads.
-
Html Report: Quality report in HTML format for immediate visualizaiton.
-
Json Report: Quality report in Json format for MultiQC.
FastQC
FastQC is a program for quality control of high-throughput sequencing datasets and exports several analyses statistics into nicely formatted HTML reports. It can identify problems in the raw sequencing data early on. Besides basic statistics, it analyzes per base sequence quality, per base N content and sequence length distribution.
Tool used: FastQC
Input
- Samples: List of prefiltered fastq RNA-seq samples.
Input Parameters
-
Min Length: (optional) Sets an artificial lower limit on the length of the sequence to be shown in the report. As long as you set this to a value greater or equal to your longest read length then this will be the sequence length used to create your read groups. This can be useful for making directly comaparable statistics from datasets with somewhat variable read lengths.
-
Dup Length: (optional) Sets a length to which the sequences will be truncated when defining them to be duplicates, affecting the duplication and overrepresented sequences plot. This can be useful if you have long reads with higher levels of miscalls, or contamination with adapter dimers containing UMI sequences.
-
Kmers: Specifies the length of Kmer to look for in the Kmer content module. Specified Kmer length must be between 2 and 10. Default length is 7 if not specified.
-
No Group: When set will disable grouping of bases for reads >50bp. All reports will show data for every base in the read. WARNING: Using this option will cause fastqc to crash and burn if you use it on really long reads, and your plots may end up a ridiculous size. You have been warned!
Output
-
HTML report: FastQCs results in HTML format.
-
Zip report: FastQCs results as compressed Zip file.
BBSplit
BBSplit is binning reads to references by mapping them to multiple references at the same time. This is useful when the input files contain reads from several different organisms as each reads will be assigned to the organism(s) it maps to best.
There are several ways of obtaining the "Index"-files, BBSplit accepts either an existing file (see Load BBsplit Index node) or a freshly generated file using the BBSplit Index Reference node and the BBsplit Build Index node.
Tool used: BBMap
BBSplit Build Index
Builds indexes that can be used for the BBSplit Index Reference node.
Takes several reference fasta files and builds an index from them. This index can then be used by the BBSplit node to map reads against all the references simultaneously.
There is always one primary reference marked by "Primary Reference".Additional reference fasta files can be supplied by using the BBSplit Index Reference node. multiple Index reference nodes can be chained together for combining multiple secondary references into one index.
The output will be a reference to a .tar.gz archive, that compresses a directory named "BBSplit". The index reference can either be directly forwarded to the BBSplit node, or can be reused by using the "Load BBSplit Index" node.
Input
-
Primary Reference: Fasta containing the primary reference for the index. Will be passed to the 'ref_primary' argument.
-
References: List of additional references. Can be obtained from the 'BBSplit Index References' node.
Input Paramters
- Index Name: Name of the BBSplit index created.
Output
-
BBSplit Index: Compressed BBSPlit index directory. An archive containing a single directory called 'BBSplit' into which the BBSplit index is written.
-
BBSplit Log: Log for BBSplit index creation.
BBSplit Index References
Aggregates a list of reference fasta files for the BBsplit Build Index.
Input
-
Fasta: File ID referring to the fasta file containing the reference sequences.
-
References: List of references to append the new reference to. If empty, a new list is created.
Input Parameters
- Name: Name (ID) of the reference created.
Output
- References: Aggregated list of references (name-file-id pairs) that can be forwarded to the BBsplit node.
Load BBSplit Index
Load an already computed BBSplit Index, saving resources by not having to compute it again each time.
Input
- Tar Gz File: BBSplit index to load in a compressed ".tar.gz" file.
Output
- BBSplit Index: Can be forwarded to the BBSplit node, so the index doesn't have to be freshly computed each time running the pipeline.
BBSplit
BBSplit maps reads to multiple references simultaneously. The references to map against are provided via an index that can be computed with the BBsplit Build Index node and loaded with the Load BBsplit Index node.
The reads are then written to a file for the reference they best match, a read can match one, all or no reference. This is done for each sample separately.
Input
-
Samples: Samples containing the reads to map to the references.
-
BBSplit Index: ".tar.gz" file that contains a BBSplit index. Can be constructed using the BBsplit Build Index node or loaded using Load BBsplit Index.
Output
-
Primary Samples: Samples containing reads that belong to the primary reference.
-
Non Primary Samples: Samples containing reads that belong to the remaining references.
-
Log Files: Log files of the process.
SortMeRNA
Filter out ribosomal RNA from metatranscriptomic data by mapping reads to a selection of four prepared databases. Selecting multiple of the available databases at the same time is possible.
Tool used: SortMeRNA
Input
- Samples: List of fastq sequencing samples to extract rRNA reads from.
Input Parameters
-
Use Default DB Ref: If checked smr_v4.3_default_db.fasta is used as a reference.
-
Use Fast DB Ref: If checked smr_v4.3_fast_db.fasta is used as a reference.
-
Use Sensitive DB Ref: If checked smr_v4.3_sensitive_db.fasta is used as a reference.
-
Use Sensitive DB RFam Seeds Ref: If checked smr_v4.3_sensitive_db_rfam_seeds.fasta is used as a reference.
Output
-
Non rRNA Samples: Updated samples, now referencing .fastq.gz files that do not contain rRNA reads.
-
Log Files: Log files of the process.
2. Alignment and Quantification
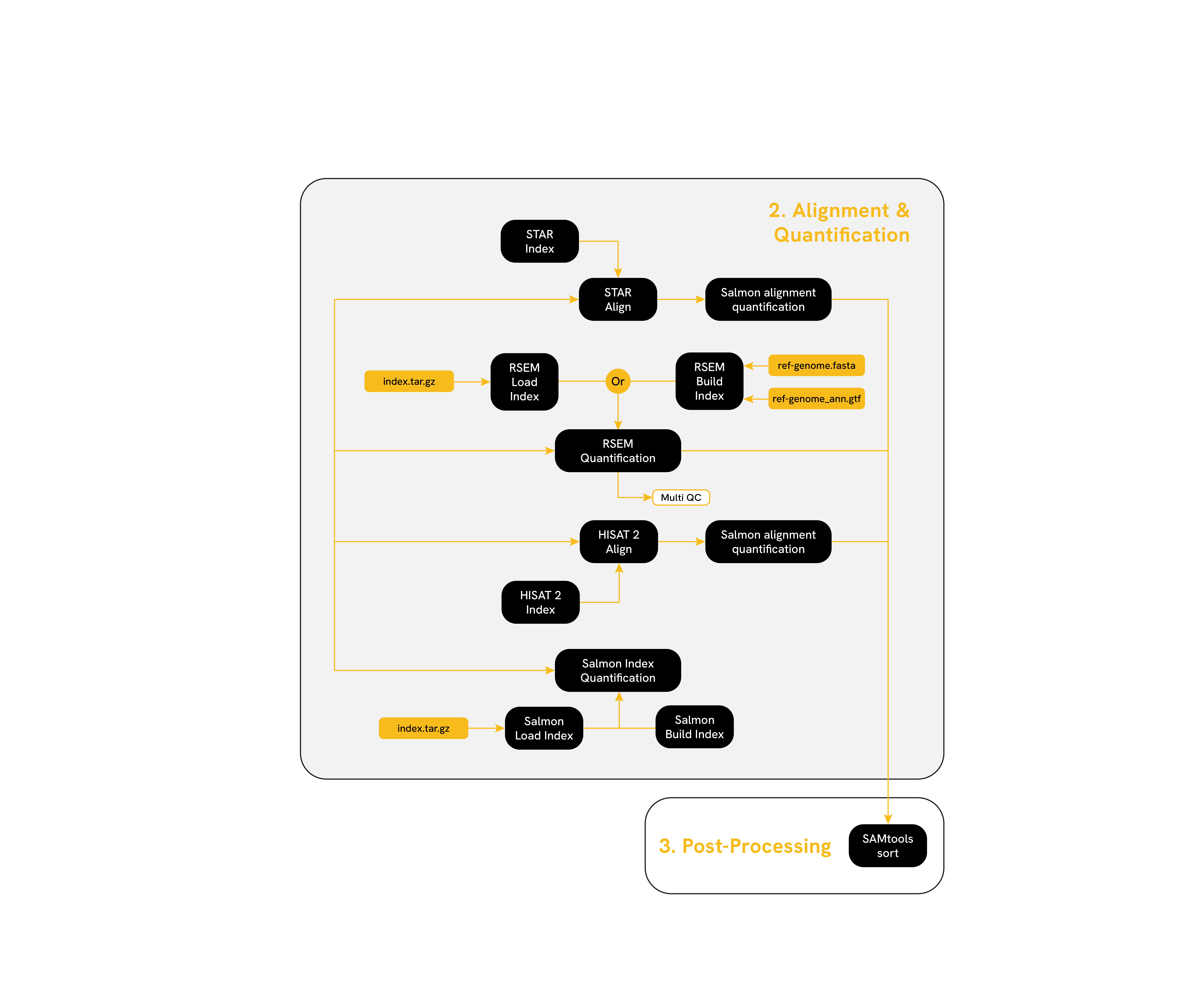
Alignment is the process of assigning each read to a specific place in the genome (or transcriptome) where it was transcribed from. This is an essential step to find out which transcripts are found in the sample and as a result which genes are being transcribed by the sequenced cells.
Quantification takes the alignment and tries to more accurately compute the expression level of each gene taking into account the distortions caused by PCR and other processing steps.
The pipeline provides many different options for alignment and quantification of the preprocessed reads. The simplest one is to use RSEM to both align and quantify reads in one node. Salmon can also be used to pseudoalign and quantify transcripts in a signle node, but while this is faster and less computationally intense it does not generate an actual alignment that can be used in further steps of the pipeline. It is also possible to only align the reads using either HISAT2 or STAR and rely on coverage results later in the pipeline entirely foregoing quantification, or to combine either alignment tool with a Salmon quantification step.
RSEM
RSEM aligns and quantifies the given samples to a reference genome in the form of a precomputed index. It generates a BAM containing the computed alignment and a quantification report. To run the RSEM Quantification Node requires an index of a reference genome matching the organism the reads were sampled from. This index can be newly generated by the RSEM Build Index node or an existing index can be loaded using RSEM Load Index.
Tool used: RSEM
RSEM Build Index
Create an RSEM index for a desired genome that can be used to align and quantify reads from that organism. The Fasta and gtf files required can be obtained from many public genome libraries, but two commonly used sources are GENCODE for human and mouse genomes and ENSEMBL for other common organisms. Be aware of what version of the genome is used as it is recommended to only use the primary asssembly for both Hisat2 and STAR alignments to avoid issues with haplotypes increasing multimapper rates.
Input:
-
Genome: The fasta file containing the genome of the reference organism.
-
Annotation: The gtf file containing the matching genome annotations.
Input Parameters:
- Aligner: The alignment program to use. Determines which aligner has to be used in the RSEM Quantification node. For more information on benefits and drawbacks of each alignment tool - see the following sections on STAR and HISAT2.
Output:
- RSEM Index: The generated genome index compressed into a tar.gz archive.
RSEM Load Index
Loads an RSEM Index present in the files section so it does not have to be newly generated each time the pipeline is executed.
Input
- Tar Gz File: RSEM index to load in a compressed ".tar.gz" file.
Output:
- RSEM Index: The selected genome index.
RSEM Quantification
Executes the alignment and quantification for the provided reads and outputs seperate alignment (BAM) and quantificaion (Sf) files.
Input:
-
Samples: Preprocessed fastq files containing the reads to be aligned.
-
RSEM Index: Genome index matching the organism the reads were obtained from.
Input Parameters:
- Aligner: The alignment program to use. Has to be the same as the aligner set when generating the genome index. For more information on benefits and drawbacks of each alignment tool - see the following sections on STAR and HISAT2.
Output:
-
Gene Quantification: File containing the quantification of approximate expression levels of the genes found in the index (Sf file).
-
Alignment: The alignment computed by the selected alignment tool. Is used downstream to analyze gene coverage and the quality of the RNA-seq results. Alignments are output in BAM format and sepertely in transcriptome and genome coordinates.
-
Logs: Log files that can be fed to MultiQC to visualize results in a report.
STAR
STAR (Spliced Transcripts Alignment to a Reference) is a high-performance alignment tool used for aligning RNA-seq reads to a reference genome or transcriptome. It is particularly effective at handling large datasets, as it is optimized for high accuracy, with capabilities to detect splice junctions and handle large RNA-seq datasets efficiently. However it is more resource intensive compared to other aligners like HISAT2 or Salmons pseudoalignment. Currently there is no option to build custom STAR indices and all indices have to be selected from a list of pregenerated references using the STAR Index node.
Tool used: STAR
STAR Index
Loads a pregenerated STAR index and a matching gtf annotation file for use in the STAR Align Node.
Output:
-
STAR index: The selected STAR index.
-
Annotation: The matching gtf genome annotation file.
STAR Align
Executes the alignment for the provided reads.
Input:
-
Samples: Preprocessed fastq files containing the reads to be aligned.
-
STAR Index: Genome index matching the organism the reads were obtained from.
Input Parameters:
-
Gene Annotation: A gtf annotation file containing information about gene locations, splice sites, and possible transcripts. This file is optional, but highly recommended as it greatly improves alignment accuracy. A gtf file fitting the pregenerated indices is output by the STAR Index node.
-
Read Length: Length of the longest read of the given samples. Setting the correct value might improve alignment accuracy, but the default option usually provides sufficient results.
-
Save Unmapped: When set the node will output a list of unmapped reads to a seperate file. Other tools can try to "rescue" these reads by performing a more intense alignment or the distribution of unmapped reads can be part of downstream analysis.
Output:
-
Alignments: List of BAM files containing the alignments in genome and transcriptome coordinates seperately.
-
Logs: List of Logs with details about each STAR run.
-
Unmapped reads: (optional) List of unmapped reads (Fastq).
HISAT2
HISAT2 is an alignment program designed for fast and accurate alignment of RNA-seq reads to a reference genome. It is particularly suited for handling large datasets and complex genomes with many known splicing events, using an efficient indexing method that supports gapped alignments and splice-aware mapping. However it does not perform as well with novel spilce sites or genomes without extensive annotation. There is also no option to output alignments in transcript coordinates, which could be quantified by Salmon. Currently there is no option to build custom HISAT2 indices and all indices have to be selected from a list of pregenerated references using the HISAT2 Index node.
Tool used: HISAT2
HISAT2 Index
Loads a pregenerated HISAT2 index for use in the HISAT2 Align node.
Output:
- HISAT2 Index: The selected HISAT2 index.
HISAT2 Align
Executes the alignment for the provided reads.
Input:
-
Samples: Preprocessed fastq files containing the reads to be aligned.
-
HISAT2 Index: Genome index matching the organism the reads were obtained from.
Input Parameters:
- Save Unmapped: When set the node will output a list of unmapped reads to a seperate file. Other tools can try to "rescue" these reads by performing a more intense alignment or the distribution of unmapped reads can be part of the downstream analysis.
Output:
-
Alignments: List SAM files containing the alignments in genome coordinates.
-
Unmapped Reads: (optional) List of unmapped reads (Fastq).
Salmon
Salmon is a software tool used for quantifying transcript abundance in RNA sequencing data. It employs a fast and accurate pseudoalignment method to estimate gene and transcript expression levels, making it efficient for bulk RNA sequencing analysis, especially in large datasets. It is often used to perform differential expression analysis, alternative splicing analysis and other downstream genomic tasks.
There are two methods for running Salmon:
-
The first is to run it directly on the preprocessed reads using the Salmon Index Quantification node. This skips the alignment step, performing a pseudoalignment instead, and quantifying based on that. Since no actual alignment was performed the reads genome coordinates are unknown making many downstream analyses impossible. Because of this only the quantification data can be used.
-
The second is to use STAR to perform a transcript level alignment and the quantify based on the alignment using the Salmon Alignmnet Quantification node. This enables more varied downstream analyses, thought it is significantly more computationally intense and as such much slower.
Tool used: Salmon
Salmon Index Quantification
This way of running Salmon skips the alignment of reads and instead performs a pseudoalignment based on a Salmon reference index generated by the Salmon Build Index node. It takes the reads and an index to directly produce a quantification file.
Input:
-
Samples: Preprocessed fastq files containing the reads to be quantified.
-
Salmon Index: Genome index matching the organism the reads were obtained from.
-
Annotation: A gtf file containing the annotation for the matching organism genome.
Output:
-
Quanfification: A list of .sf files containing the quantification results.
-
Log archives: A list of archives that can be passed to MultiQC to visualize results in a report.
Salmon Build Index
Generates an index required for running the index based quantification of Salmon. This is not required when working with already aligned reads in the Salmon Alignment Quantification node.
The Fasta and gtf files required can be obtained from many public genome libraries, but two commonly used sources are GENCODE for human and mouse genomes and ENSEMBL for other common organisms. Be aware of what version of the genome is used as it is recommended to only use the primary asssembly for both Hisat2 and STAR alignments to avoid issues with haplotypes increasing multimapper rates.
Input:
-
Genome: The fasta file containing the genome of the reference organism.
-
Transcriptome: The fasta file containing the matching transcriptome (This can be generated from the genome and a gtf annotation file).
Output:
- Salmon Index: The generated genome index compressed into a tar.gz archive.
Salmon Load Index
Loads a Salmon Index present in the files section so it does not have to be newly generated each time the pipeline is executed.
Input
- Tar Gz File: Salmon index to load in a compressed ".tar.gz" file.
Output:
- Salmon Index: The selected genome index.
Salmon Alignment Quantification
This version of running Salmon requires already aligned reads that were aligned to the transcriptome coordinates. This can only done using STAR with the Align to Transcript option enabled. This node does not require a Salmon genome index to work.
Input:
-
BAM Files: Files contianing the aligned reads. No metadata required.
-
Transcriptome: Transcriptome of the organism in question. The names of the transcripts here have to match the names of the transcripts used in the alignment.
Output:
-
Quanfification: A list of .sf files containing the quantification results.
-
Log archives: A list of archives that can be passed to MultiQC to visualize results in a report.
3. Post-Processing
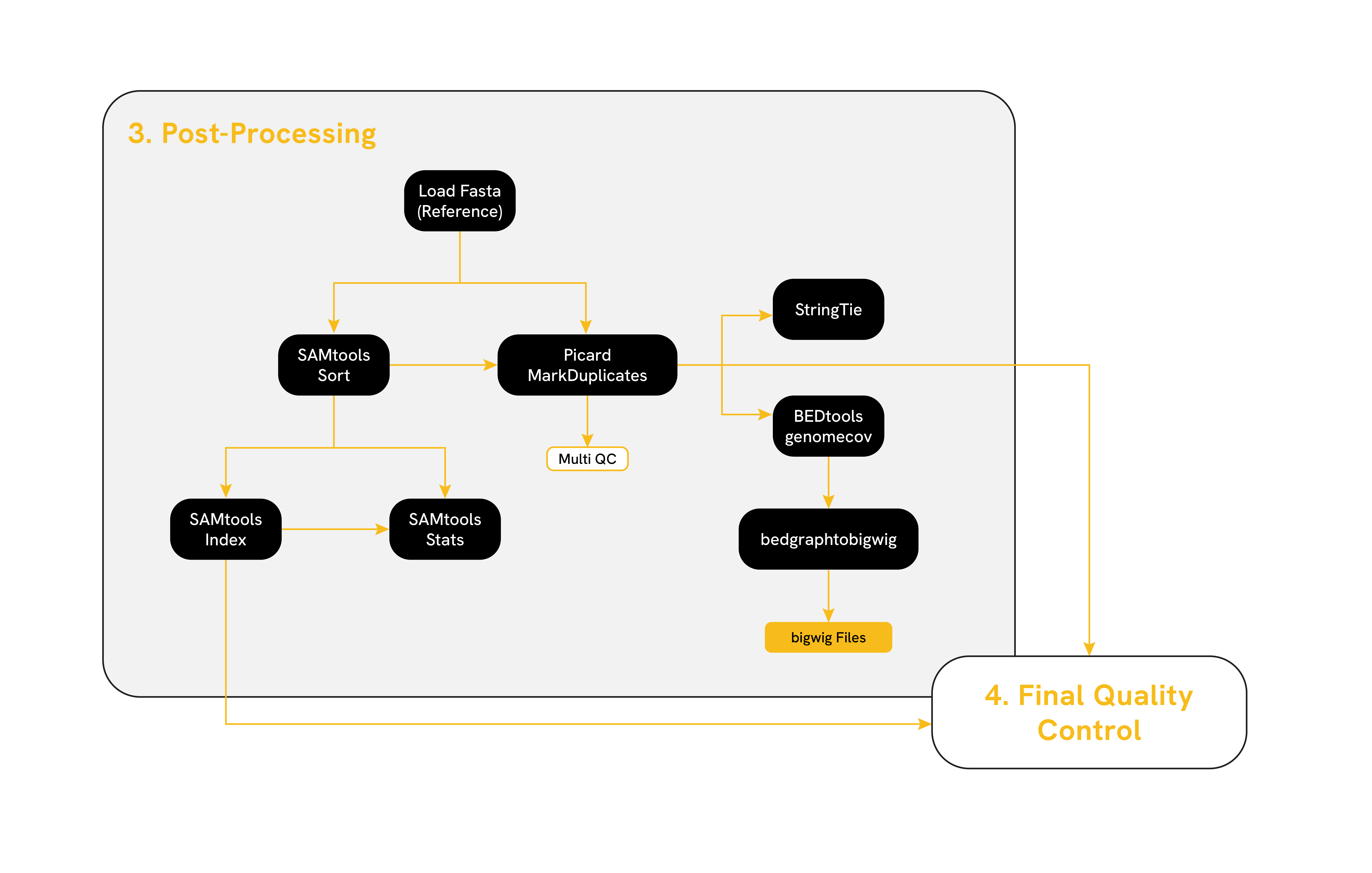
The postprocessing steps ensure accurate transcript quantification and further refinement of RNA-seq data. These steps include alignment sorting and indexing with (SAMtools), removal of PCR duplicates using (Picard MarkDuplicates), assessment of genome coverage with (BEDtools genomecov), and transcript assembly and quantification with (StringTie). These tools collectively enable enhanced accuracy in downstream analyses, such as differential expression or transcript isoform identification.
SAMtools
SAMtools is a collection of tools for working with SAM and BAM alignment files. Many downstream analysis steps require the alignments to be either sorted, indexed or converted from SAM to BAM and SAMtools can execute all these operations quickly. The different functions of SAMtools are seperated into their own nodes so the operations for a specific pipeline can be used independently.
Tool used SAMtools
SAMtools sort
The sort node will sort alignments in a given BAM file by their genome (or transcriptome) coordinates. A reference can be supplied in which case entries will be sorted by their coordinates in the reference instead. Specifying a reference will cause the sorted BAM to not be compatible with the SAMtools index node where it will behave like an unsorted BAM. The output BAM file will keep the input BAMs metadata.
Input:
- Bam Files: List of alignments to be sorted.
Input Parameters:
- Reference: (optional) Fasta file containing a reference for an alternative sort order.
Output
- Sorted Bams: List of sorted alignments.
SAMtools index
Creates an index for fast random access of alignments in the corredponding BAM file.
Input:
- Bam Files: List of sorted alignments.
Input Parameters:
- Index Type: Type of index to generate (bai or csi).
Output:
- Bam Indices: List of alignment indices (BAI/CSI).
SAMtools stats
Provides summary statistics including basic information such as the number of reads, sequence lengths, and mapping quality. It also reports on alignment metrics like coverage, insert size distribution, and errors or discrepancies between the reference and aligned reads. For a more detailed breakdown of all the gathered statistics see here. The generated stats file can be fed to MultiQC to be included in a full report.
Input:
- Bam Files: List of alignments to summarize.
Input Parameters:
- Reference: Provide a reference genome file to compute additional statistics (GC-depth and mismatches-per-cycle).
Output:
- Statistics: List of gathered statistics that can be passed to MultiQC.
Picard MarkDuplicates
Detects duplicate reads in a provided BAM file. Detected duplicates can either be tagged appropriately in the file for further processing or be removed entirely. The node will also generate a text file with duplicate statistics that can be fed to MultiQC for a combined report. The input has to contain strandedness metadata and must be sorted.
Tool used: Picard
Input:
-
Bam Files: List of sorted alignments to check for duplicates.
-
Reference: (optional) A fasta file containing a reference sequence. This is optional but will increase duplicate mapping accuracy.
Input Parameters:
-
Sort Order: The node will assume the input files are sorted according to this criterion. If unknown is set it will try to find the sort order in the file header.
-
Remove Duplicates: If set the node will remove duplicates instead of only marking them.
-
Validation Stringency: Set the validation stringency. Setting stringency to SILENT can improve performance when processing a BAM file in which variable-length data (read, qualities, tags) do not otherwise need to be decoded.
Output:
-
Marked Bams: Alignments with duplicates marked or removed.
-
Metrics: List of duplicate metrics that can be passed to MultiQC.
BEDtools genomecov
Computes bedgraph reports of aligned sequences for the given BAM files. The bedgraph will show the coverage of the reads continuously across the whole genome and can be used to visualize read depth across the genome. The input has to contain strandedness metadata and must be sorted.
Tool used: BEDTools
Input:
- Bam Files: List of sorted alignment files to generate genome coverage from.
Input Parameters:
-
Strand: For which strand to generate the coverage graph (forward or reverse).
-
Sort: If set the node will group the lines by chromosome and sort them by their position.
Output:
- Bedgraph Files: List of coverage graphs.
bedgraphtobigwig
Converts Bedgraph files to BigWig format. Unlike bedGraph, bigWig allows efficient visualization and retrieval of specific genomic regions without loading the entire dataset, making it more scalable for large sequencing data.
Tool used: USCS bedgraphtobigwig
Input:
-
Bedgraph files: List of Bedgraph files to convert.
-
Chromosome size: Files describing the chomosome sizes. The file has to contain two-columns: | chromosome name || size in bases |.
Input Parameters:
- Fileextension Marker: (optional) Addition to the file extension. Name of the generated files will be Sample.marker.bigwig.
Output:
- BigWig Files: List of converted files.
StringTie
StringTie is a transcript assembler used to reconstruct full-length transcripts, estimate gene and transcript expression levels, and identify novel isoforms. StringTie is particularly useful for differential expression analysis, transcript discovery, and annotation refinement, often working in conjunction with reference annotations and featureCounts or Ballgown for downstream analysis. The input BAM files must be sorted.
Input:
- Bam Files: Aligned reads in BAM format. Alignment has to be sorted by genomic coordinates (Output by the samtools sort node).
Input Parameters:
- Reference Annotation: With a reference .gtf file a file containing covered transcripts and a ballgown table can be generated.
Output:
-
Potential Transcripts: Transcripts assembled by stringtie (Gtf).
-
Quantification: Estimates for the assembled transcripts expression level (Txt).
-
Covered Transcripts: (optional) Annotation containing only transcripts that were fully covered by reads (Gtf),
-
Ballgown Table: (optional) Files usable like those produced by the 'tablemaker' program included with the Ballgown distribution.
4. Final Quality Control
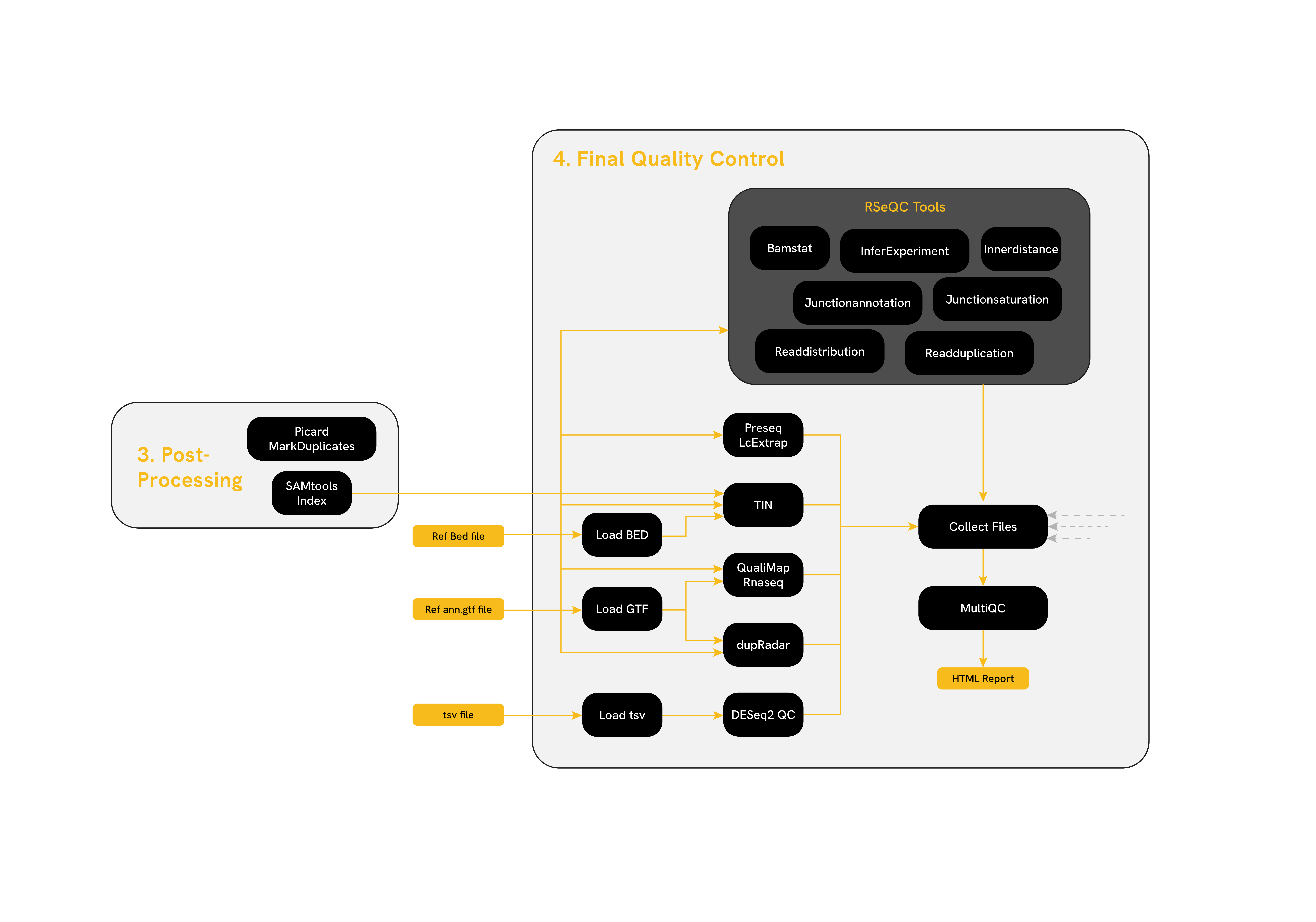
The final quality control provides further quality metrics and combines all previously calculated QCs into one extensive report. Additional metrics include evaluation of read distribution and strand-specific bias (RSeQC), prediction of read depth saturation (Preseq LcExtrap), quality control of read alignments (Qualimap Rnaseq), accession of duplication rates (dupRadar) and PCA differential gene expression analysis (DESeq2 QC).
RSeQC
RSeQC is a quality control toolset for RNA-seq data that evaluates various metrics to assess sequencing quality, alignment performance, and potential biases. It analyzes features like read distribution, gene body coverage, GC content, junction saturation, and duplication rates, helping identify issues such as rRNA contamination or incomplete gene coverage. There are many different possible statistics that can be generated by seperate nodes that are useful for different applications. The generated statistics can be passed to MultiQC for a full report.
Bamstat
Bamstat provides a quick summary of alignment quality from a BAM file. It reports key metrics helping assess the overall quality of RNA-seq or genomic alignments. This tool is useful for most pipelines, regardless of what further processing will be done, by helping detect issues like excessive multi-mapping or low mapping rates before proceeding with downstream analysis.
Input:
- Bam Files: BAM files containing read alignments of the samples (need normal meta data; i.e. strandedness info is not required).
Output:
- Statistics: Txt report containing summary of calculated statistics that can be passed to MultiQC.
InferExperiment
InferExperiment determines the library strandness of an RNA-seq dataset by analyzing how reads align to the reference genome. If already stranded alignments are provided the node will recheck if the strandedness is correct and summarize the results in a .tsv file that can also be passed to MultiQC.
Input:
-
Bam Files: BAM files containing read alignments of the samples (need normal meta data; i.e. strandedness info is not required).
-
Reference Bed: Bed file for the reference gene model.
Input Parameters:
-
Fail Strand Check Filename: Filename for the strand check report.
-
Stranded Threshold: The portion of reads that have to match a certain strandedness to classify the reads as a whole as that strandedness.
-
Unstranded Threshold: The portion of reads that have different strandedness for the reads to be classified as unstranded.
Output:
-
Library Strandedness: Txt report summarizing the nodes analysis that can be passed to MultiQC.
-
Fail Strand Check: TSV report indicating if the inferred strandedness matches the provided one, or the one inferred by Salmon. Can be passed to MultiQC.
Innerdistance
Innerdistance calculates the inner mate distance (or insert size) between paired-end RNA-seq reads. It measures the distance between the 3′ end of the upstream read and the 5′ end of the downstream read when mapped to the same transcript. This is only applicable for paired end alignments and helps detect potential issues like fragmentation bias or incorrect pairing. The results will be saved to multiple txt files that can be fed to MultiQC to generate a final report as well as a pdf containing a seperate graphical summary.
Input:
-
Bam Files: BAM files containing read alignments of the samples (need normal meta data; i.e. strandedness info is not required).
-
Reference Bed: Bed file for the reference gene model.
Output:
-
Distance: Paired end distance, frequencies and means reports that can be passed to MultiQC (Txt).
-
Plots: Graphical summary of distances (Pdf).
-
R script: Script used to generate plot.
Junctionannotation
Junctionannotation analyzes splice junctions from RNA-seq data to assess splicing accuracy and detect novel or misannotated junctions. It compares detected splice junctions against a reference annotation (GTF file) and classifies them into categories from known to unannotated junction. The node generates multiple reports about the junctions and plots showing the different splice events of each gene.
Input:
-
Bam Files: BAM files containing read alignments of the samples (need normal meta data; i.e. strandedness info is not required).
-
Reference Bed: Bed file for the reference gene model.
Output:
-
Junction Information: Xls files containing junction information.
-
Bed Files: Junction and Interact Bed files.
-
Plots: junction and Event plots (Pdf).
-
R script: Script used to generate plot.
Junctionsaturation
Junctionsaturation evaluates the sequencing depth sufficiency for detecting splice junctions in RNA-seq data. It does this by randomly subsampling reads at different depths and checking how many junctions can still be detected, helping assess whether sequencing depth is adequate for capturing splicing events. This is particularly important for alternative splicing analysis, as a sequencing run that does not reach saturation with known junctions is unlikely to produce accurate results for new splicing events. The node produces a plot showing if a sample reached saturation in known, novel and all junctions.
Input:
-
Bam Files: BAM files containing read alignments of the samples (need normal meta data; i.e. strandedness info is not required).
-
Reference Bed: Bed file for the reference gene model.
Output:
-
Saturation Plot: Plot showing junction detection at various subsampling percentages (Pdf).
-
R script: Script used to generate plot.
Readdistribution
Readdistribution analyzes how RNA-seq reads are distributed across different genomic features, such as exons, introns, UTRs, intergenic regions, and coding sequences (CDS). This helps assess library quality and potential biases, such as excessive intronic or intergenic reads, and is a good quality estimator for almost all downstream analyses. The generated statistics can be passed to MultiQC for a summary report.
Input:
-
Bam Files: BAM files containing read alignments of the samples (need normal meta data; i.e. strandedness info is not required).
-
Reference Bed: Bed file for the reference gene model.
Output:
- Read Distribution: Text file conatining calculated distibution statistics that can be passed to MultiQC.
Readduplication
Readduplication assesses PCR and optical duplicate levels in bulk RNA-seq data by analyzing the redundancy of reads based on their mapping position and sequence. High duplication rates indicate bias and may suggest poor library complexity, helping detect overamplification during library preparation. The results will be saved to seperate Xls files that can be fed to MultiQC to generate a final report as well as a pdf containing a seperate graphical summary.
Input:
- Bam Files: BAM files containing read alignments of the samples (need normal meta data; i.e. strandedness info is not required).
Output:
-
Duplication Rate: Duplication rate statistics determined seperately by position and sequence (Xls).
-
Duplication Plot: Plots for visualization (Pdf).
-
R script: Script used to generate plots.
TIN
TIN calculates the Transcript Integrity Number (TIN) for each alignment providing a transcript-level assessment of RNA integrity. TIN scores range from 0 to 100, with higher scores indicating more uniform read coverage across a transcript. The median TIN across all transcripts in a sample serves as an overall indicator of RNA quality, aiding in the detection of RNA degradation which is crucial for accurate downstream analyses. The statistic summary Txt file can be passed to MultiQC.
Input:
-
Bam Files: BAM files containing read alignments of the samples (need normal meta data; i.e. strandedness info is not required).
-
Bam Indices: The BAM indices for the provided BAM files. Also need normal meta data to find the correct BAM-BAI pairs. A bam index is paired with a BAM file, if the sample name in the sample meta data is the same.
-
Reference Bed: Bed file for the reference gene model.
Output:
-
TIN: Xls File containing the TIN for each gene.
-
Summary: Text file containing summary statistics for eacht input BAM, which can be passed to MultiQC.
Preseq LcExtrap
LcExtrap estimates library complexity and sequencing saturation by extrapolating the number of unique reads expected at increasing sequencing depths. It helps determine if additional sequencing would yield more unique reads or just more duplicates. The generated Txt report can be passed to MultiQC. Preseq is prone to fail often with low complexity or small files and while there are some options that can mitigate this behavior it may be best to skip this node if it fails on a particular dataset.
Tool used: Preseq
Input:
- Bam Files: BAM files containing read alignments of the samples (need normal meta data; i.e. strandedness info is not required).
Input Parameters:
-
No Defects: If set preseq will not test for defects. May help if the run fails without known cause.
-
Segment Length: Maximum segment length when mergin paired end bam.
-
Retry Without Pe: If set the node will try to rerun preseq without the paired end flag set as this flag can often cause preseq to fail.
-
Seed: (optional) Sets a seed for the random generator.
Output:
-
Library Complexity Extrapolation: Output generated by preseq lc_extrap (can be passed to MultiQC). Contains info on total number of reads, average expected number of distinct reads, lower and upper limit of the confidence interval.
-
Logs: Log files of the process.
Qualimap Rnaseq
Qualimap is a tool for assessing the quality of read alignments. The RNA-seq mode is used and supplies metrics likereads genomic origin, junction analysis, transcript coverage and 5'-3' bias computation.
Tool used: Qualimap
Input:
-
Bam Files: BAM files containing the alignments of your samples, stranded meta-data required.
-
Genome Gtf: Annotations of your reference genome (Ensembl GTF file format).
Output:
- Reports: Archived html quality reports and statistics (can be passed to MultiQC).
dupRadar
Assesses duplication rates (PCR artifacts) in RNA-Seq datasets graphically.
Tool used: dupRadar
Input:
-
Bam Files: BAM files containing the alignments of your samples, stranded meta-data required.
-
Genome Gtf: Annotations of your reference genome (Ensembl GTF file format).
Input Parameters
- Feature Type: Describes which features from the GTF file should be considered when calculating the duplication metrics (default: exon).
Output:
-
Scatter 2D: Plot containing the duplication rate against the total read count.
-
Boxplot: Duplication rate vs. total reads per kilobase (RPK) boxplot.
-
Hist: Expression histogram showing reads per kilobase (RPK) values per gene.
-
Dupmatrix: Text file containing tags falling on the features described in the GTF file.
-
Intercept & Slope: Text file containing intercept and slope from dupRadar modelling.
-
MultiQC: dupRadar files to pass to MultiQC.
DESeq2 QC (PCA)
Performs PCA on the gene counts for each sample.
The main input is a .tsv file with a row for each gene and a column for each sample (and a gene_name column) indicating how often a gene was represented in each sample. The makes each sample to an N-dimensional data point where N is the number of genes covered in the .tsv file.
| gene_id | gene_name | sample1 | sample2 | ... |
|---|---|---|---|---|
| 1 | GeneA | 4 | 2 | |
| 2 | GeneB | 5 | 6 | |
| ... |
The PCA then projects the N-dimensional data points into a 2D plane in such a way that as much information as possible is preserved. The results for that PCA are then returned in the form of plots, parameter files and other reports.
Tool used: DESeq2
Input:
- Counts: A tsv file with the following columns:
gene_id gene_name sample1 sample2 ...where the sample columns contain count values. Can be obtained from the TxImport node.
Input Parameters
-
Label: Label to add to headers of output tsv files. This is so Multiqc can properly label the sections in that report. E.g., the sections will then be named '
-
Use Vst: Run 'vst transform' instead of 'rlog' for stabilizing the count data. vst is overall faster, whereas rlog is more robust in cases with highly variable counts and library size. For further information about the two options, consult the DESeq2 manual here.
Output:
-
Plots: A PDF containing PCA plots and a heatmap for the distances between samples.
-
RData: R DESeqDataSet object that stores 'read accounts and intermediate estimated quantities during statistical analysis'.
-
PCA Txt: Text file that stores the 2D coordinates of the samples obtained from the PCA.
-
PCA MultiQC File: Same as the 'PCA Txt' output but prepended with a header containing meta data for MultiQC.
-
Dists: Text file that stores the sample to sample distances obtained from the PCA.
-
Dists MultiQC File: Same as the 'Dists' output but prepended with a header containing meta data for MultiQC.
-
Log: File containing R session info. E.g., the used R version, the used packages, ...
-
Size Factors: A .tar.gz archive of the generated 'size_factors' directory. The directory contains a .txt file for each sample containing a factor such that all the count vectors by all samples can be brought to a common scale by dividing by the corresponding size factor. It also contains a .RData file containing the size factors vector as a whole.
MultiQC
Collect Files
A node for collecting all reports created along the pipeline and aggregating them to one report file that can be submitted to the MultiQC node.
Input
-
Files A: First file to aggregate with the second file. Can be either a file or a file list.
-
Files B: Second file.
Output
- Combined Files: Combined file list. The metadata of each file is conserved.
MultiQC
MultiQC aggregates results from previous analyses across many samples into a single report.
You can use the Collect Files node, to aggregate all those files and use them as input for this node.
Tool used: MultiQC
Input
- Report Files: Already aggregated report files (use Collect Files for file aggregation).
Input Parameter
- Report Name: Name of the MultiQC file report.
Output
- HTML Report: Combined report containing all reports that have been input.